Association Mapping: An Important Tool for Crop Improvement
Mitali Tiwari
July 2nd 2025, 12:21:52 pm | 5 min read

Association mapping seeks to identify specific functional genetic variants linked to phenotypic differences in a trait to facilitate detection of trait-causing DNA sequence polymorphisms and selection of genotypes that closely resemble the phenotype.
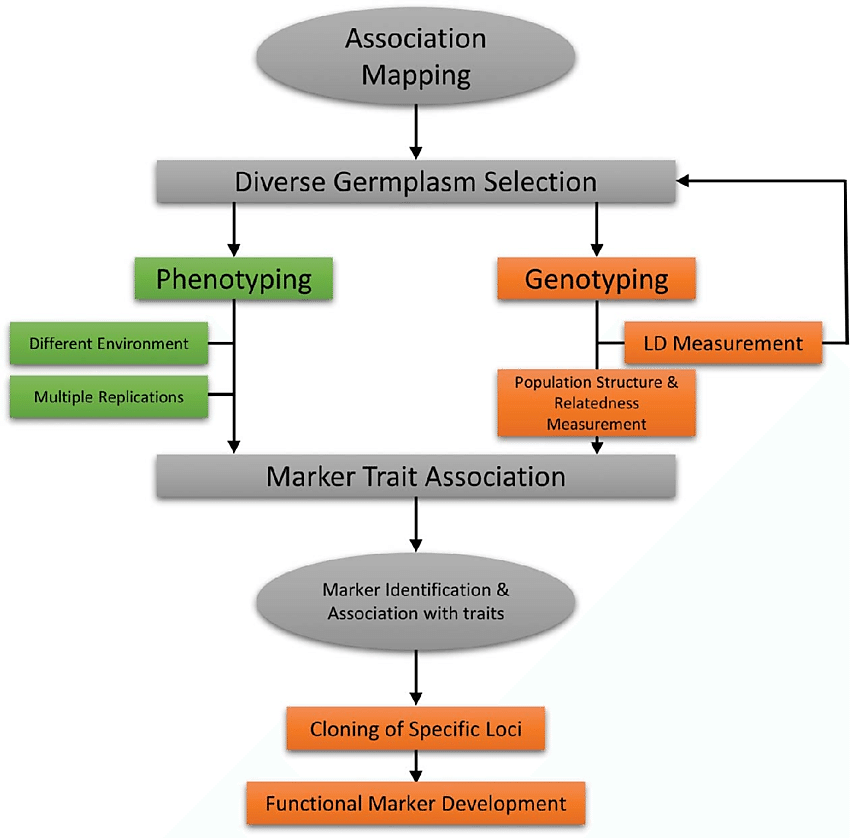
Figure: Methodology of association mapping (Source: Nadeem et al., 2018)
- The primary purpose of gene mapping is to take advantage of molecular markers that are closely related to the genes that regulate quantitative features, enabling marker-assisted selection.
- Linkage analysis and linkage disequilibrium based on association mapping are the two techniques most frequently used for breaking down complex features.
- Linkage analysis has inadequate marker resolution due to the minimal number of recombination events that take place during the formation of mapping populations at the expense of propagating and evaluating a large number of lines.
- A population-based study called association mapping, commonly referred to as LD mapping, employs linkage disequilibrium to discover associations between trait markers.
- Association mapping has evolved as a method for resolving complex trait variation down to the sequence level by exploiting historical and evolutionary recombination events at the population level.
Association mapping offers three advantages over traditional linkage analysis.
- Higher mapping resolution
- Less research time
- A larger number of alleles.
Linkage Disequilibrium (LD)
Robbins introduced LD for the first time in 1918. The non-random relationship between alleles of two or more loci is referred to as LD.
Association mapping involves the following steps:
(1) Individual lines are randomly selected from breeding populations, germplasm collections, or wild populations.
(2) Exact phenotyping across time or environments.
(3) Marker-based genotyping, in which a sufficient number of molecular markers are examined. SSR markers are preferably used (genotyping).
(4) The STRUCTURE and TASSEL (Trait Analysis by Association, Evolution, and Linkage) programs must be used to determine the population structure and kinships or relatedness, respectively.
(5) Various statistical techniques are employed to quantify connection.
Fig. Analysis through STRUCTURE and TASSEL software
Two approaches under association mapping:-
1. Candidate gene association mapping
2. Genome-wide association mapping.
1. Candidate-gene association mapping: The candidate gene technique is a hypothesis-driven strategy for dissecting complicated traits with the goal of locating the most significant alleles. This procedure entails genotyping or resequencing the genes within the test germplasm that are thought to have a high probability of correlation with the desired trait. Although this approach is costly, it may find rare polymorphisms that the first method could have missed. The benefit of resequencing the complete gene is that it can quickly determine which haplotype is appropriate for each breeding objective.
2. Genome-wide association mapping: A thorough technique is taken to thoroughly explore their genome for unintentional genetic variation. Without the need for prior knowledge of potential genes, many markers are evaluated for associations with a variety of complicated phenotypes. A research consortium with complementary skills and sufficient money operates most effectively.
Advantages of association mapping-
- An effective method for finding new genes or quantitative trait loci (QTLs) for significant agronomic traits is association mapping.
- When predicting phenotype, gene-based markers are more reliable than linked markers.
- The best haplotype across one or more genes can be predicted using the results of association studies for the best expression of the desired trait.
- Human genetics researchers are actively using genome-wide association analyses to map disease-causing genes.
- It offers mapping with a high resolution (less than 10 centimorgan).
- Each locus can have more than two alleles being researched at once.
- Less time spent conducting research to create an association.
Factors affecting association mapping:
1. Population structure
2. Small sample size
3. Low frequency of specific alleles that may increase the detection of a false positive associations.
4. Genetic drift
5. Natural and artificial selection
6. Mating system and admixture of different populations
Advanced approaches:
Multiparent advanced generation intercross (MAGIC): Mott et al. initially suggested this technique in mice. Through the use of various combinations of numerous parents, it is possible to identify the genes responsible for the quantitative qualities. Steps in this includes:
- Founder selection
- Mixing of parents
- Advanced intercrossing
- Inbreeding
Thus, it has the potential to increase the speed and efficiency of breeding
Nested association mapping (NAM): For high resolution and high power mapping of complex traits, it is an integrated multi-parent population technique that combines the benefits of linkage mapping and association mapping.
Conclusion:
Association mapping offers great potential to enhance crop genetic improvement superior alleles and to provide a large number of lines. The power of AM is to detect many superior alleles while association mapping is influenced by allele frequency distribution at the functional polymorphism level. In this regard, several studies have combined linkage mapping and LD mapping, a methodology known as Nested association mapping which reduces spurious associations caused by population structure. Still extensive research is needed to understand the expansive application of association mapping.
References:
Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. science. 2008 Nov 7; 322(5903):881-8.
Kushwaha UK, Mangal V, Bairwa AK, Adhikari S, Ahmed T, Bhat P, Yadav A, Dhaka N, Prajapati DR, Gaur A, Tamta R. Association mapping, principles and techniques. J. Biol. Environ. Eng. 2017;2(1):1-9.
Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, Hatipoğlu R, Ahmad F, Alsaleh A, Labhane N, Özkan H. DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnology & Biotechnological Equipment. 2018 Mar 4;32(2):261-85.
Zhu C, Gore M, Buckler ES, Yu J. Status and prospects of association mapping in plants. The plant genome. 2008 Jul;1(1).