PRE-BREEDING - Harnessing the power of plant genetic resources
Nethala Aneesha, Rumana Khan, and Akhouri Nishant Bhanu
June 27th 2025, 6:07:40 pm | 5 min read

Introduction
Crop domestication and improvement can be seen as a series of rounds of selection that eventually isolates agriculturally beneficial genetic variety from ancestor’s wild species. The crops that feed the world today were created through these iterative rounds of selection, but at the expense of their genetic diversity, leaving them with less allelic diversity than their wild progenitors and other crop wild relatives (Meena et al., 2017).
Natural genetic variation within crop species has been used since the dawn of agriculture to provide enough food for subsistence, and today efforts are being made to produce enough food in excess to feed an expanding population. The so-called genetic bottleneck, which occurs in wild populations, causes drastic population declines, a loss of genetic diversity, and increased susceptibility to infectious diseases and pests, which increases the likelihood that the particular crop in question would go extinct (Govindaraj et al., 2015). Due to biased breeding techniques that focused on improving only a few traits (like yield and its component traits), frequent use of a small number of carefully chosen genotypes as parents in varietal development programmes, and introduction of a small number of outstanding lines to many countries, natural variability was depleted, which increased genetic similarity between modern crop cultivars. Further crop variety improvement will be challenging given the decreased genetic diversity (Bhandari et al., 2017). Pre-breeding, or genetic enhancement, has only recently become a required, regular, and planned component of all plant breeding activities, as well as a crucial component of germplasm diversification schemes. Rick (1984) coined the term "pre-breeding" (Rao et al., 2013). The most effective method of tying breeding projects and genetic resources together is pre-breeding. It refers to all methods used to isolate desired traits and/or genes from un-adapted (exotic or semi-exotic) materials, including those that have undergone any type of selection for improvement while being adapted. It is a crucial component of initiatives for germplasm diversification. To introduce new, superior features that aren't present in the indigenous cultivars, genetic enhancement is used. The materials generated during pre-breeding are anticipated to be suitable for inclusion in regular breeding procedures (Iqbal et al., 2013).
The most frequent breeding techniques used to increase production are selection or hybridization, which expand the gene pool with local or foreign material. The set of all genes in any population, often of a specific species, is known as the gene pool. All cultivars, wild species, and wild relatives that possess all the genes useful for breeding are included in the crop's gene pool (Majhi, 2020).
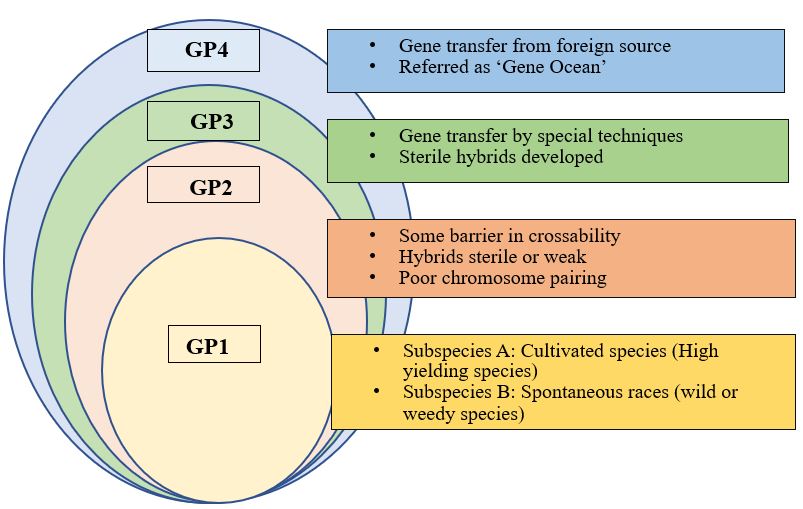
Fig: Modified gene pool concept in plants based on the hybridization study (a revised form of Harlan and De Wet, 1971)
Pre-breeding uses a larger pool of genetic resources to create a new base population for breeding programmes (Haussmann et al., 2004). It has been used successfully in several crops (rice, tomato, soybean, cotton, maize, wheat, barley, groundnut, chickpea, pigeon pea, sorghum, pearl millet) and improved many cultivated varieties for various qualitative and quantitative traits. Pre-Breeding operations have been started in a variety of programmes using potential landraces, wild relatives, and well-known cultivars (Jain & Prakash, 2019). Broadening the base of gene banks and pre-breeding that relate to the conservation and uses of plant genetic resources (PGR) is required to close the gap between those plant breeders working with advanced and elite cultivars and those using wild genetic resources. The effective administration of the gene bank, both ex-situ and in situ, and the identification of the genes in the gene bank through gene discovery are prerequisites for pre-breeding (Surya, 2017). The awareness of CWRs' value for food security and the advancement of modern biotechnologies have led to recent advances in the usage of wild resources.
The examples in other studies (Brozynska et al., 2015; Colmer et al., 2006; Ford Lloyd et al., 2011; Hajjar & Hodgkin, 2007; Maxted & Kell, 2009; Nevo & Chen, 2010; Zamir, 2001) show that a wealth of genetic diversity has been preserved in wild relatives of different crops, much of which has yet to be explored.
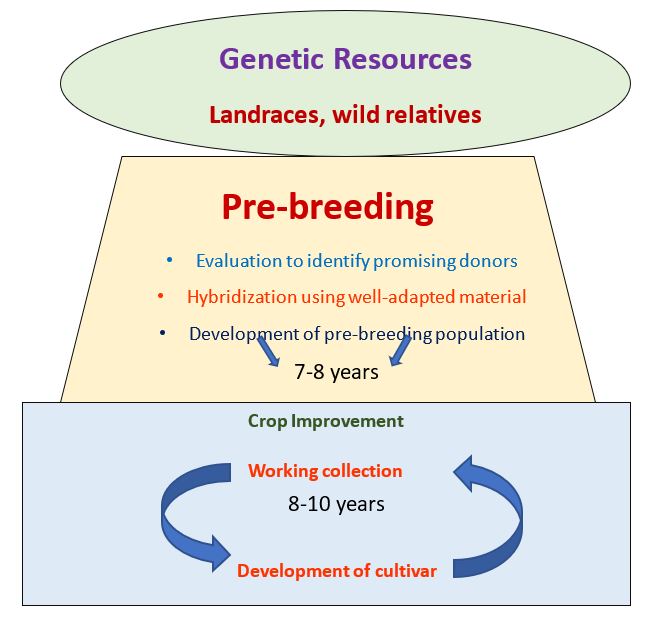
Fig: Pre-breeding as a bridge between genetic resources and crop improvement
(Sharma et al. 2013)
Objectives of Pre-breeding:
The following four goals are addressed during pre-breeding work: -
1) Using a larger pool of genetic material that limits genetic uniformity in crops, 2) identifying desirable traits/genes and transferring them 3) developing parental stocks that are easily used in breeding programs and 4) finding potentially useful genes in a well-organized and documented gene bank (Abebe & Tafa, 2021).
Pre-breeding is decided upon based on the anticipated effectiveness and efficiency of transferring the intended features into cultivars for farmers and the source of the desired gene(s) in the long run. If desired genes are only present in one of the following, pre-breeding is required:
1) Gene bank accessions that are poorly adapted to the target environment, 2) closely related wild species that are easily crossed with crop species and 3) more distant wild species that are more challenging to cross with crop species (Kumar, 2018).
Methods in pre-breeding:
In pre-breeding, there are two separate methods—introgression and incorporation—for employing wild species or foreign material (Simmonds, 1993).
Introgression: It denotes the transfer of one or a few alleles from exotic genotypes to breeding populations that have been acclimated but do not have the alleles that regulate a particular attribute. Improvement via introgression is often the most affordable and quick way to improve the chosen pre-breeding accessions when better germplasm is already available from ongoing breeding programmes (Akkenapally & Shiva Kumar, 2022).
1) Recurrent backcross: where the donor parent and recurrent parent are crossed frequently, allowing for the easy transmission of highly heritable monogenic or oligogenic traits.
2) Inbred backcross: It requires one to three backcrosses. Traits from exotic germplasm are successfully transferred into elite adapted genetic populations.
3) Congruity backcross: Both the donor and the recurrent parent are backcrossed using this technique in alternating generations.
Incorporation: Refers to increasing the genetic diversity of new breeding materials by creating locally adapted populations (genotypes) utilising alien germplasm. Some genetic principles of integration were published by Loknathan et al., (2003) including the utilisation of unadapted material and material with a large range of variability. evaluating adaptation in various agroclimatic situations. The entire method works in conjunction with traditional breeding and, by using unadapted stocks, leads to the formation of future parents (Pathak & Srivastava, 2021).
The adoption of crop wild relatives (CWRs) is a viable strategy to promote the genetic diversity of cultivated crops since the limited genetic basis of current cultivars is becoming a significant bottleneck for crop improvement efforts. Due to the significant degree of linkage drag brought on by the CWR, the introduction of desirable characteristics from a CWR to a sexually compatible conventional line through traditional breeding may provide additional challenges. But marker-assisted backcrossing has been demonstrated to be a method capable of quickly getting rid of linkage drag with the fewest possible generations (Mammadov et al., 2018).
Limitations of pre-breeding:
1) Longer time required (5 to 10 years more) 2) Access to and exchange of germplasm is difficult due to intellectual property rights 3) New types of production practise emerge 4) Cross incompatibility through wide hybridization 5) Linkage drag 6) Low levels of recombination in the hybrids 7) New pest or disease problems develop 8) Needs establish new market demands and Climate shock problems (Ponnaiah & Gunasekaran, 2020).
Techniques for alien gene transfer (Qureshi et al., 2014):
- In vitro methods and embryo rescue.
- Including bridge species that can be crossed between the two parental species.
- The use of exogenous immunosuppressants and plant growth regulators at the post-pollination stage.
- Backcrossing the hybrids with the agronomically acceptable base as the recurrent parent.
- Chromosome duplication and backcrossing when genome nonhomology (4n or 6n pathway) is present.
- Chromosome procedures like irradiation, homoeologous pairing, substitutions, alien addition lines (MAAL and DAAL), translocations, and alien addition lines to promote gene transfer.
Genes transferred for disease and insect resistance in Rice:
| Species | Gene transferred |
| O. australiensis | BLB, BPH |
| O. brachyantha | BLB |
| O. latifolia | BLB, BPH, WBPH |
| O. minuta | Blast, BLB, BPH |
| O. officinalis | BPH, BLB, WBPH, GLH |
| O. nivara | Grassy stunt |
| O. perennis | CMS |
Disease resistance genes and their source in Wheat
| Genes | Source |
| Sr 21, Sr 22 | Triticum monococcum |
| Sr 2, Sr 9e, Sr 13 | T. dicoccum |
| Sr 14, Sr 17, Sr 33 | Ae. Squarrosa |
Gene Introgression through chromosome substitution method:
|
| Resistance |
| Triticum aestivum × Haynaldia | Powdery mildew |
| Trifolium repens × T. nigrescens | Cyst nematode |
| Beta vulgaris × Brassica. species. | Nematode |
| Cicer arietinum × C. reticulatum | Cyst nematode |
| Glycine max × G. tomentella (homoeologous) | Cyst nematode |
Technologies that Can Assist or Speed-Up Pre-breeding:
(a) Trait screening methods: There is so much more complexity in the way some traits can be detected using a normal manual selection method, which is very important for increasing wheat yield potential. Pre-breeding of these characteristics can be facilitated by the development of gene markers, prediction models and approaches to the selection of genes.
(b) Genetic markers: In the case of simple traits, where genetic markers explain a significant amount of variability, they are very useful in facilitating selection and back-crossing. But understanding the genetic structure of trait, heritability estimate and complexity is also aided by studies on RUE for radiation efficiency and BM biomass.
(c) Gene editing: In cases where the causal variants have been established, it may be useful to use this method for simple traits. It is capable of creating new variations for testing or to produce the desired variant. It might not be possible to do so for complex traits, as only a small change in the causative genes could have an effect.
(d) Speed breeding: Rapid generation advancement may contribute to the hike of pre-breeding populations in two distinct areas before breeding: (1) development of RIL or BC populations to study the genetics of traits and (2) bulking.
(e) Reference genomes: This helps identify and cross-check new alleles. It's also useful for comparing different marker systems based on physical location. It also helps predict candidate genes for further study, cloning, and haplotyping studies.
(f) Gene cloning: Gene cloning is not required at the pre-breeding stage, but cloned genes can be anchored in elite strains and help identify new genes. (Sukumaran et al., 2022)
Various activities are involved in Pre-breeding:
I. Characterization of landrace populations:
Landraces are always heterogeneous and are an excellent source of genetic variation in plant breeding programs. These carry useful genes such as genes for early maturity, yield potential, disease and pest resistance, and other desirable traits.
II. Creation of new parent populations:
Parents with high specific or general combinatorial skills are selected by well-designed recombination progeny testing. Hence breeders continuously select potential parent populations from a variety of sources.
III. Introgression of new traits from other useful sources:
New cultivars are developed by introducing new genes into an existing genetic background.
IV. Creation of novel traits:
Induced mutagenesis through the use of artificial mutagens is an important tool for increasing mutation frequency and thereby expanding genetic diversity in plant breeding and functional genomics.
V. Creation of polyploidy:
Individuals with an altered set of chromosomes (euploids) arise either by duplicating the genome of one species or by mating unrelated species and duplicating the chromosomes of interspecific hybrids. It can be artificial by various means, such as exposing the plant material to environmental factors (cold or hot treatments, X-ray irradiation, etc.) or using chemicals that disrupt normal chromosome divisions (colchicine, etc.). can be induced.
VI. Acquisition of new information on crop genetics:
Breeders are constantly looking for new genes from various sources to increase nutritional value, early maturity, high yield potential, and resistance to biotic and abiotic stress. Understanding the candidate genes and their inheritance patterns in controlling these traits is critical for effective introduction and improved selection efficiency in cultivar development.
VII. Development of new plant breeding techniques:
New modern breeding techniques help improve selection responses. These include the development of more efficient conventional selection methods, biotechnology, molecular marker technology, and the identification of markers associated with traits of interest, potent gameticides, and cytoplasmic sterility systems with desirable genetic backgrounds.
VIII. Cultivar development:
The breeder's goal should be to strike a balance between farmer-preferred traits and solutions to production constraints to facilitate farmer dissemination of the variety. A plant breeding unit with sufficient resources needs to run two parallel programs simultaneously. First, a pre-breeding program to continuously develop new and improved parent strains with superior traits of commercial importance.
IX. Applications of doubled haploids in plant breeding:
In vitro production of haploid plants followed by somatic chromosomal duplication is the fastest means of generating pure replicated haploids (DH).
X. Pre-breeding and wide crosses:
This includes basic research to achieve wide-ranging hybridization and activities to promote the use of alien species and wild relatives. The main goal is to provide breeders with a more usable and more "attractive" PGR. (Tefera, 2021)
Pre-breeding work done in other crops using wild species:
Through inter-specific hybridization, valuable features of the soybean have been found in wild species and introduced to cultivated ones (Sebolt et al., 2000). More gene flow between domesticated and wild species has been found in maize, leading to a highly polymorphic genome, the significance of which has not yet been investigated (Roy & Wang, 1999). Due to the limited genetic diversity of cultivated kinds, over 20,000 wild rice accessions are kept in seed banks, and ongoing work is being done to introduce desired features into cultivated types (Plucknett & Smith, 2014). An essential gene was extracted from the wild tomato species (Lycopersicom pennellii B.), and it enhanced the content of pro-vitamin A in the fruit by more than 15 times (Ronen et al., 2000).
Groundnut, pigeonpea, chickpea, sorghum, and pearl millet were the five crops for which pre-breeding work was intensively conducted utilising wild species and improved cultivars (ICRISAT SS, 2004). The pre-breeding study was also conducted on several crops, including rice, wheat, sugarcane, groundnuts, tomatoes, sunflowers, cotton, and common beans (Seetharam, 2007). It was effective in introducing genes from wild species into cultivated species for certain features. Some of them are listed below:
| Crop
| Wild Relatives
| Traits (Abiotic Stress)
| Traits (Biotic Stress)
| References |
| Rice | Oryza minuta Oryza rufipogon Oryza australiensis Porteresia coarctata Oryza meridionalis O. australiensis
| Salt tolerance, heat and cold tolerance & flooding tolerance
| Blast resistance, bacterial leaf streak resistance, bacterial blight resistance, fungal diseases resistance.
| (Majee et al., 2004) (Sengupta & Majumder 2010) (Rohiniet al., 2014) (Baruahet al., 2009); (Niroulaet al., 2012); (Liu, Lu, Zeng, & Wang, 2002) |
| Barley | Hordeum spontaneum Hordeum chilense
| Drought tolerance, salt tolerance
| Fusarium crown rot resistance, scald resistance, powdery mildew/leaf rust resistance.
| (Diab et al.,2004) (Shavrukov et al., 2010) (Bahieldin et al., 2015) (Danan, Veyrieras, & Lefebvre, 2011)
|
| Wheat
| Triticum dicoccoides Triticum aegilops Triticum tauschii Triticum monococcum Agropyron elongatum Aegilops species | Drought tolerance, salt tolerance, O3 tolerance.
| Powdery mildew fungus resistance, stem rust resistance, leaf rust resistance
| (Di Bianco et al., 2011); (James et al.,2012); (Xue, Ji, Wang, Zhang, & Yang, 2012); (Periyannan et al., 2013) |
| Peanut
| Arachis stenosperma Arachis duranensis Arachis ipaënsis
| Drought and fungal resistance
| Nematode resistance
| (Guimaraes et al., 2012); (Chu, Holbrook, Timper, & Ozias Akins, 2007) |
| Soybean
| Glycine soja Glycine tomentella
| Salt tolerance Drought tolerance
| Nematode resistance
| (Ha et al., 2013); (Luo et al., 2013); (Winter, Shelp, Anderson, Welacky, & Rajcan, 2007) |
Future Prospects:
- Due to the increased probability of extinction for endemic and narrowly adapted species, there is an urgent need for the collection, characterisation, and documenting of wild species, especially crop wild relatives.
- The necessity for effective screening of germplasm for various features such, as quality traits and biofortification, as well as the requirement for adapting agriculture to biotic and abiotic challenges, has raised the demand for new genes in germplasm/genebank collections.
- The wheat and barley genome mapping and synteny of the gene sequenced can be allocated to encoding abiotic stress tolerance and used for crop development.
- To transfer the desired gene(s) from the tertiary gene pool and/or beyond, the potential of genetic transformation techniques might be used.
- To make use of the data collected from genetic tests, new breeding techniques and bioinformatics tools must be developed.
Conclusion:
Wild species are the storehouse of many useful genes and can be used effectively to generate genetic variability following wide hybridization. Successfully introducing new genetic variation to the elite cultivar with minimal linkage drag and no alteration of gene balance for an elite genotype is a crucial step in pre-breeding. There is an urgent need to strengthen pre-breeding programmes, which will ensure that new genetic variability is continuously supplied into the main breeding programmes to accelerate genetic gains and improve the nutrition and resilience of modern crop varieties. This is especially true in the era of climate change, which makes it even more important to harness the full potential of wild species. Breeding can take some time, but in particular, when there is a need for discovery and translation research as well as possible broad crossover with wild relatives or selective selection of progeny it is important to make the most of the crop's biological potential.
References:
Abebe, A., & Tafa, Z. (2021). Pre-Breeding Concept and Role in Crop Improvement. International Journal for Research in Applied Sciences and Biotechnology, 8(2), Article 2.
Akkenapally, J., & Shiva Kumar, G. (2022). Pre-Breeding: A Review. 2320–2882.
Bahieldin, A., Atef, A., Sabir, J. S. M., Gadalla, N. O., Edris, S., Alzohairy, A. M., ... Jansen, R. K. (2015). RNA-Seq analysis of the wild barley (H. spontaneum) leaf transcriptome under salt stress. Comptes Rendus Biologies, 338, 285–297.
Baruah, A. R., Ishigo-Oka, N., Adachi, M., Oguma, Y., Tokizono, Y., Onishi, K., & Sano, Y. (2009). Cold tolerance at the early growth stage in wild and cultivated rice. Euphytica, 165, 459–470.
Brozynska, M., Furtado, A., & Henry, R. J. (2015). Genomics of crop wild relatives: Expanding the gene pool for crop improvement. Plant Biotechnology Journal, 14, 1070–1085.
Chu, Y., Holbrook, C. C., Timper, P., & Ozias-Akins, P. (2007). Development of a PCR-based molecular marker to select for nematode resistance in peanuts. Crop Science, 47, 841–847.
Colmer, T. D., Flowers, T. J., & Munns, R. (2006). Use of wild relatives to improve salt tolerance in wheat. Journal of Experimental Botany, 57, 1059–1078.
Di Bianco, D., Thiyagarajan, K., Latini, A., Cantale, C., Felici, F., & Galeffi, P. (2011). Exploring the genetic diversity of the DRF1 gene in durum wheat and its wild relatives. Plant Genetic Resources, 9, 247–250.
Diab, A. A., Teulat-Merah, B., This, D., Ozturk, N. Z., Benscher, D., & Sorrells, M. E. (2004). Identification of drought-inducible genes and differentially expressed sequence tags in barley. Theoretical and Applied Genetics, 109, 1417–1425.
Ford-Lloyd, B. V., Schmidt, M., Armstrong, S. J., Barazani, O., Engels, J., Hadas, R., ... Maxted, N. (2011). Crop wild relatives-undervalued, underutilized and under threat? BioScience, 61, 559–565.
Govindaraj, M., Vetriventhan, M., & Srinivasan, M. (2015). Importance of Genetic Diversity Assessment in Crop Plants and Its Recent Advances: An Overview of Its Analytical Perspectives. Genetics Research International, 2015, 431487.
Guimaraes, P. M., Brasileiro, A. C., Morgante, C. V., Martins, A. C., Pappas, G., Silva, O. B. Jr, ... Moretzsohn, M. C. (2012). Global transcriptome analysis of two wild relatives of peanut under drought and fungi infection. BMC Genomics, 13, 387.
Hajjar, R., & Hodgkin, T. (2007). The use of wild relatives in crop improvement: A survey of developments over the last 20 years. Euphytica, 156, 1–13.
Haussmann, B. I. G., Parzies, H. K., Presterl, T., Sui, Z., & Miedaner, T. (2004). Plant genetic resources in crop improvement. Plant Genetic Resources: Characterization and Utilization, 2(1), 3–21.
ICRISAT, International Crop Research Institute for semi-arid tropics 2004. Success story 22 Jan 2004 Success story SS 22 – Jan 2004. Patancheru, Hyderabad, India. online website. www. Icrisat.org
Iqbal, A. M., Lone, A. A., Wani, S. A., Wani, S. H., & Nehvi, F. (2013). Pre-breeding and Population Improvement. LS: International Journal of Life Sciences, 2(3), 188.
Jain, S. K., & prakash, O. (2019). Pre-breeding: A Bridge between Genetic Resources and Crop Improvement. International Journal of Current Microbiology and Applied Sciences, 8(02), 1998–2007.
James, R. A., Blake, C., Zwart, A. B., Hare, R. A., Rathjen, A. J., & Munns, R. (2012). Impact of ancestral wheat sodium exclusion genes Nax1 and Nax2 on grain yield of durum wheat on saline soils. Functional Plant Biology, 39, 609–618.
Kumar, V., & Shukla, Y. M. (2014). Pre-breeding: Its applications in crop improvement. Double Helix Res, 16, 199-202.
Liu, G., Lu, G., Zeng, L., & Wang, G. L. (2002). Two broad-spectrum blast resistance genes, Pi9(t) and Pi2(t), are physically linked on rice chromosome 6. Molecular Genetics and Genomics, 267, 472–480.
Loknathan, T. R., Singh, P., Agarwal, D. K., Mohan, P., Singh, S. B., Gotmare, V., & Singh, V. V. (2003). GENETIC ENHANCEMENT IN COTTON.
Majee, M., Maitra, S., Dastidar, K. G., Pattnaik, S., Chatterjee, A., Hait, N. C., ... Majumder, A. L. (2004). A novel salt-tolerant L-myo- inositol-1phosphate synthase from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice—molecular cloning, bacterial overexpression, characterization, and functional introgression into tobacco-conferring salt tolerance phenotype. Journal of Biological Chemistry, 279, 28539–28552.
Majhi, P. K. (2020). Chapter-4 Gene pool concept in plant breeding. Current research and innovations in plant pathology, 85.
Mammadov, J., Buyyarapu, R., Guttikonda, S. K., Parliament, K., Abdurakhmonov, I. Y., & Kumpatla, S. P. (2018). Wild Relatives of Maize, Rice, Cotton, and Soybean: Treasure Troves for Tolerance to Biotic and Abiotic Stresses. Frontiers in Plant Science, 9, 886.
Maxted, N., & Kell, S. P. (2009). Establishment of a global network for the in-situ conservation of crop wild relatives: Status and needs. Rome, Italy: Food and Agriculture Organization of the United Nations Commission on Genetic Resources for Food and Agriculture.
Meena, A., Gurjar, D., & Kumhar, B. (2017). Chemical Science Review and Letters Pre-breeding is a Bridge between Wild Species and Improved Genotypes-a Review.
Nevo, E., & Chen, G. X. (2010). Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell and Environment, 33, 670–685.
Niroula, R. K., Pucciariello, C., Ho, V. T., Novi, G., Fukao, T., & Perata, P. (2012). SUB1A-dependent and -independent mechanisms are involved in the flooding tolerance of wild rice species. The Plant Journal, 72, 282–293.
Pathak, S. K., & Srivastava, N. PRE-BREEDING: IT’S APPLICATION IN CROP IMPROVEMENT ARTICLE ID.: 11.
Periyannan, S., Moore, J., Ayliffe, M., Bansal, U., Wang, X. J., Huang, L., ... Lagudah, E. (2013). The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science, 341, 786–788.
Plucknett, D. L., & Smith, N. J. H. (2014). Gene Banks and the World’s Food. Princeton University Press.
Ponnaiah, D. G., & Gunasekaran, D. K. (2020). Pre-Breeding and Its Importance in Crop Improvement Program. 1, 3.
Qureshi, A., Lone, A., Wani, A., Wani, S., & Nehvi, F. (2014). Pre-breeding and Population Improvement. LS-International Journal of Life Sciences, 2, 188–197.
Rao, D. M. R., Jhansilakshmi, K., Saraswathi, P., Rao, A. A., Ramesh, S. R., Borpuzari, M. M., & Manjula, A. (2013). Scope of pre-breeding in mulberry crop improvement—A review. 1(6).
Rohini, G., Mohit, V., Shashank, A., Rama, S., Manoj, M., & Mukesh, J. (2014). Deep transcriptome sequencing of wild halophyte rice, Porteresia coarctata, provides novel insights into the salinity and submergence tolerance factors. DNA Research, 21, 69–84.
Ronen, G., Carmel-Goren, L., Zamir, D., & Hirschberg, J. (2000). An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proceedings of the National Academy of Sciences, 97(20), 11102–11107.
Roy, J.D., Kindigar, B., Sinclair, T.R. 1999. Introgressing root parenchyma, Info. Maize, Maydica 44, 113-117
Sebolt, A. M., Shoemaker, R. C., & Diers, B. W. (2000). Analysis of a Quantitative Trait Locus Allele from Wild Soybean That Increases Seed Protein Concentration in Soybean. Crop Science, 40(5), 1438–1444.
Seetharam, A. (2007). Pre-breeding: An important step in the effective utilization of conserved germplasm. National Workshop on Utilization of Wild Mulberry Genetic Resources 2nd & 3rd Nov, 9–16.
Sengupta, S., & Majumder, A. L. (2010). Porteresia coarctata (Roxb.) Tateoka, a wild rice: A potential model for studying salt-stress biology in rice. Plant Cell and Environment, 33, 526–542.
Shavrukov, Y., Gupta, N. K., Miyazaki, J., Baho, M. N., Chalmers, K. J., Tester, M. ,... Collins, N. C. (2010). HvNax3-a locus controlling shoot sodium exclusion derived from wild barley (Hordeum vulgare ssp. spontaneum). Functional & Integrative Genomics, 10, 277–291.
Sukumaran, S., Rebetzke, G., Mackay, I., Bentley, A. R., & Reynolds, M. P. (2022). Pre-breeding Strategies. In M. P. Reynolds & H.-J. Braun (Eds.), Wheat Improvement: Food Security in a Changing Climate (pp. 451–469). Springer International Publishing.
Surya, M. I. (2017). Pre-Breeding and Gene Discovery for Food and Renewable Energy Security.
Xue, F., Ji, W. Q., Wang, C. Y., Zhang, H., & Yang, B. J. (2012). High density mapping and marker development for the powdery mildew resistance gene PmAS846 derived from wild emmer wheat (Triticum turgidum var. dicoccoides). Theoretical and Applied Genetics, 124, 1549–1560.
Zamir, D. (2001). Improving plant breeding with exotic genetic libraries. Nature Reviews Genetics, 2, 983–989.